
2023 was another successful year, bringing awards, progress in important research areas, and a new director to PSC! We’d like to highlight just some of the newsworthy accomplishments that the year brought.

J. Barr von Oehsen, our new director
Leadership
We were very happy to welcome our new director, Barr von Oehsen, in May. Barr joined us from Rutgers University in New Jersey.
In an early interview, von Oehsen said, “As someone who has a strong connection to the Cyberinfrastructure (CI) Ecosystem, I am very familiar with PSC and its many achievements and reputation as a global leader. The thought of being involved with moving PSC to the next level in advancing science and research is extremely exciting and an honor.”
And we’re excited to see where his leadership takes us in the years to come.
Research
As always, researchers made significant accomplishments in 2023 thanks to PSC resources.
How do human cultures and beliefs change through time? A team from Carnegie Mellon University and the Santa Fe Institute used a mathematical approach that compared data from 407 world religions as if they existed on a “landscape” where functional patterns of worship and belief are found at local “peaks,” possibly separated from each other by wide and hard-to-cross “valleys.” Their work on our Bridges-2 system showed that the approach can explain how some religions persist, others change, and still others die out, offering a new tool for understanding how human traditions develop and diverge. Read more in Cultural “Landscapes” Uses Metaphor of Valleys, Mountains, and Plains to Understand the Evolution of World Religions
G protein coupled receptors and G proteins are the targets for one third to one half of all drug therapies, directly or indirectly. Researchers from the University of California, Davis used Anton 2, designed and developed by D. E. Shaw Research (DESRES), and hosted at PSC, to simulate a G protein and its interactions with a cell-surface beta-adrenergic receptor protein known to trigger its activity. These simulations revealed movements in both molecules that had not been seen before. The results offer clues to designing drugs to better target G protein activity to treat specific diseases. Read more in “Invisible” Protein Movements Revealed by Anton 2 Simulations.
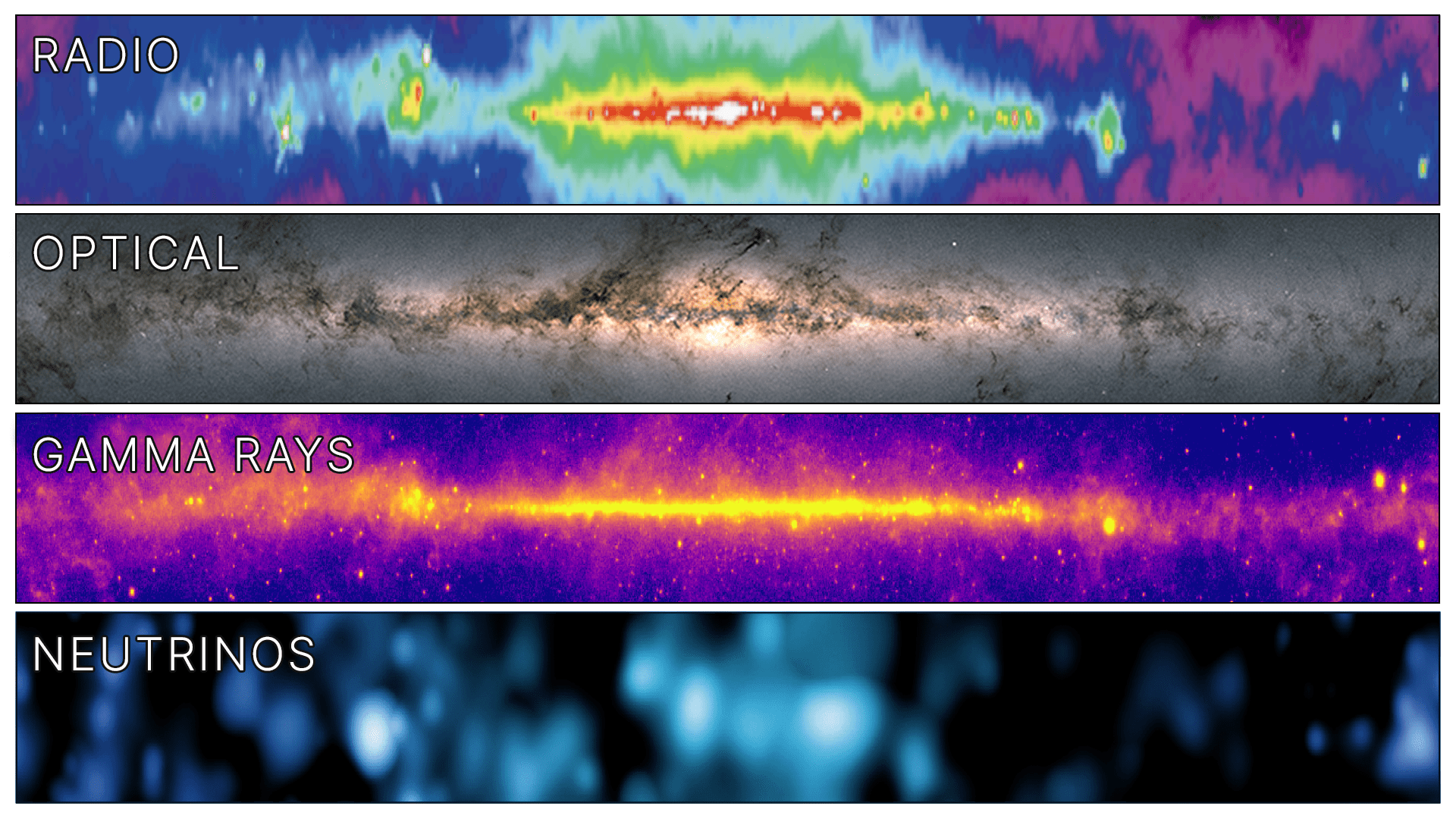
For a long time, we have observed the Universe using electromagnetic waves, whether visible light or radio waves, but those electromagnetic waves can be distorted, absorbed, and generally scrambled by stuff in between us and what we are looking at. Using our Bridges-2 system to simulate signals in their Antarctic IceCube detector, an international collaboration of scientists has now made the first map of the Milky Way galaxy using particles called neutrinos — the first map of a cosmic structure that didn’t depend on electromagnetic waves. Read more in IceCube Observatory Creates First Map of Milky Way Without Using Electromagnetic Waves.
It sounds counterintuitive, but growing more food does not necessarily translate to feeding more people. A team led by Public Health Computational, Informatics, and Operations Research, at the time based at the Johns Hopkins Bloomberg School of Public Health and currently based at City University of New York, used the HERMES supply chain software developed by scientists there and at PSC to model vegetable distribution in Odisha, a state in India. Their findings show that growing more vegetables wouldn’t increase the supply if the delivery network can’t move the veggies before they spoil. The initial study is a successful proof-of-concept for using HERMES, previously employed to simulate vaccine and other medical supply chains, to study food availability. Read more in Increasing Vegetable Crops Won’t Ease Hunger if Supply Chains Don’t Keep Pace.
Neurotransmitters are the body’s chemical messengers, helping us to move, feel sensations, and helping to keep our hearts beating, and the amino acid glutamate is the chief excitatory neurotransmitter in vertebrates. Unfortunately, neurotransmitter over-activity following a stroke or brain injury can kill neurons and cause lasting damage. A team from Johns Hopkins University School of Medicine used the Anton 2 supercomputer to simulate D-serine interacting with a key brain cell-surface protein, and found that, at high enough levels, D-serine can change from strengthening glutamate signals to blocking them. The unexpected discovery, confirmed by lab experiments, opens the possibility of future treatments to halt nerve cell overactivity. Read more in Anton 2 Simulations Reveal Unexpected Role for Brain Neurotransmitter.

The Nature cover image. Illustration credit Heidi Schlehlein, Indiana University, and the HuBMAP Consortium.
Biomedical projects
We’re proud to be part of several NIH-funded projects which are advancing biomedical research nationwide.
The Brain Image Library team was represented at the annual Society for Neuroscience Meeting 2023 in Washington, DC, where they shared new developments including a new plugin for napari designed specifically for visualizing BIL data, and the BIL Image Explorer that allows full-resolution browsing of large 3D images over the web.
The HuBMAP consortium added more than 620 datasets on their publicly available data portal, thanks in part to the hard work of our team. In July, the consortium published a series of papers in Nature, which give insight into the progress of the initiative thus far.
Outreach
We continue to provide training and education to the national community, and are proud of strides we’ve made this year to expand these efforts while also bringing them closer to home. Through the new ADAPT PA program, we’re providing curriculum development support and computing resources to community colleges and state schools across Pennsylvania.
We also worked with 13 interns this summer and are so grateful for their contributions and teamwork at PSC. You can read some of their stories in our summer wrap up post. We are especially proud that one of our interns was able to leverage his research experience here to secure a full-time job! New internship opportunities will be posted early in the new year – keep an eye on our careers page for the announcement.
Awards
Congratulations go out to our data curation dream team, Brendan Honick, Mariah Kenney, and Jackie Uranic, who won the Best Faculty/Staff Poster award at Gateways 2023 for their poster, “Data Curation Toolbox Empowering FAIR Data Sharing Via Science Gateways”.
We were honored to receive two HPCwire awards at SC23. In addition to the Editor’s Choice Award for Best HPC in Industry discussed above we won the Reader’s Choice award for Best Use of HPC in Energy for work done by a multi-institutional team which discovered how a coal-like material can be converted to a graphitic form, which is vital for clean energy and other advanced applications, including lithium ion batteries. Read more about that research in Ohio University Simulations on PSC Supercomputer Transform Coal-Like Material to Amorphous Graphite and Nanotubes.

Mariah Kenney, data curator and metadata librarian
Staff
We are incredibly grateful for our staff, because they make the magic happen. We highlight just some of the contributions that staff make in our Inside PSC series. This year, for example, we featured Chris Rapier and his HPN-SSH work and the work of our metadata librarians, among others.
None of the research that happens at PSC happens in a vacuum. Our Advanced Systems and Operations and Networking teams work in the background every day, making sure that the computing resources and networks we depend on stay up and up-to-date. The Advanced Systems group expanded and supported not only our publicly available systems, but clusters for the college of engineering and the McWilliams Center for Cosmology at Carnegie Mellon. Networking staff just completed a major network upgrade that will have data humming along for years to come. And the folks in the Support for Scientific Applications group not only installed, managed and updated the software on our computing resources, but answered user questions — over 2000 in 2023.
We were delighted to add six new staff members in 2023 in addition to Barr. Xiao, Holly, Valerie, Riaz, Dana and Jim are already making a difference here – welcome to you all.
