bridges & bridges-2 Power discovery of two-x-chromosome male voles
The Way This Rodent Species Determines Sex Differs from All Other Mammals Known
by Ken Chiacchia
Sex can be complicated. In creeping voles, a small rodent in the Pacific Northwest, it’s even more so. Scientists using PSC’s Bridges and Bridges-2 advanced research computers to assemble DNA sequences discovered that, rather than the XX sex chromosomes that determine genetic females and XY that determine genetic males in other mammals, this species has been doing just fine with only a single X chromosome in females and a strange combination of an X and another, different X in males. The work raises important questions on how early sexual development works.
Why It’s Important
In nature, sex can be really complicated. Earthworms are both sexes at once, and impregnate each other when they mate. When a queen bee mates, her offspring will all be female. If she doesn’t mate, they’ll all be male. And some female lizards can reproduce by parthenogenesis. Without needing to mate, they can produce female offspring.
The situation varies less among mammals—mostly. Over 150 million years ago, when mammals arose, they “settled” on the X and Y sex chromosome arrangement we all learned in school. A mammalian egg usually only contains a single X chromosome, because genetic females are XX. It merges with a sperm—which can contain either an X or a Y chromosome, since genetic males are XY. The resulting combination of the single sex chromosome each reproductive cell carries will determine the genetic sex of the offspring, XX or XY.
The creeping vole, also known as the Oregon meadow mouse, has turned this regularity on its head. When it arose roughly 1.2 million years ago, this Pacific Northwest rodent made a huge departure from mammalian evolution that sets them apart from even closely related voles. Scientists have known since the 1950s that creeping vole females and males weren’t quite XX/XY. In 1963, the famous geneticist Susumo Ohno proposed that creeping vole females were “XO,” having only a single X chromosome, while males were XY. But Ohno died before he could untangle what was really going on.
“This is a puzzle that started in the 1960s with an incredibly famous geneticist, Ohno. This is a system that he originally really thought was incredibly interesting, but he and his contemporaries were not able to finish. And we were able to finish that with this last puzzle piece…I think if I could have told him the story…he would have been really happy.”—Brian Couger, Brigham and Women’s Hospital.
A collaboration of scientists at Harvard Medical School-affiliated Brigham and Women’s Hospital; San Francisco State University; the University of California, Merced; and elsewhere, decided to complete Ohno’s work using the latest tools in genetic sequencing and computational assembly. Employing PSC’s National Science Foundation-funded Bridges and Bridges-2 advanced research computing platforms, Brigham and Women’s Brian Couger led the computational part of the work. He leveraged the high-memory power of those systems to assemble and identify the genes on the vole’s sex chromosomes. What the collaborators found was even more surprising than Ohno had guessed—surprising enough to earn the team a cover article in the prestigious journal Science.
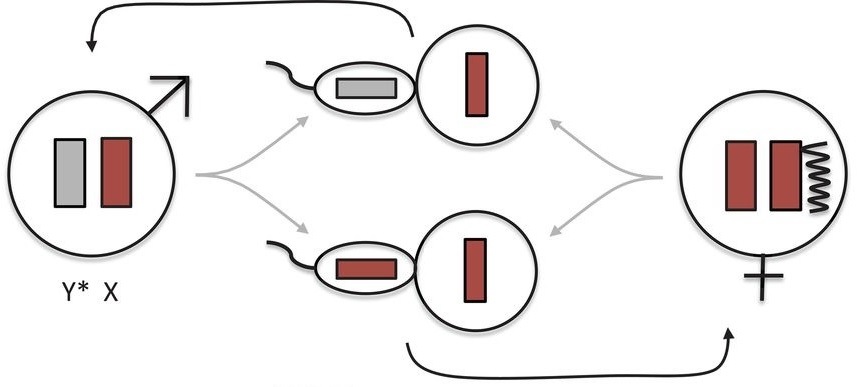
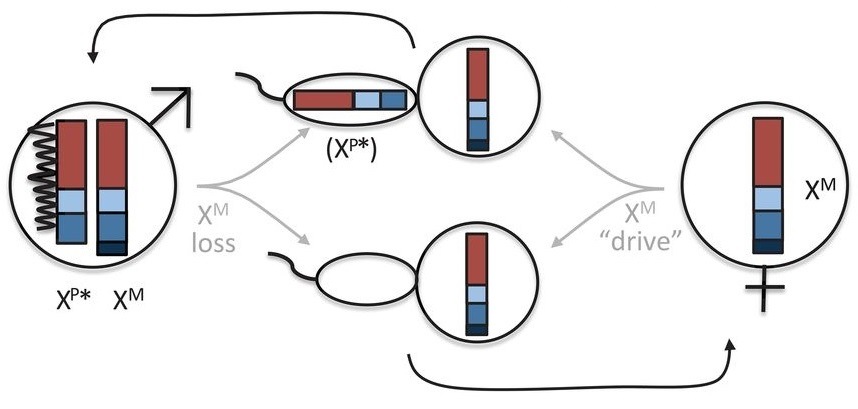
In most mammals (top), the father (left) contributes an X or a Y chromosome and the mother (right) two Xs. When the sperm and egg merge, XY makes a genetic male offspring, XX female (center). To avoid double-strength X genes, in females one X chromosome is silenced (wiggly line). Leaping voles turn this convention on its head (bottom), with the father contributing only an XP chromosome. A second, XM, chromosome is mostly or all silenced in most of the male’s cells, again shown by a wiggly line. That XM is lost when sperm cells form (center). The female in turn only contributes XM chromosomes (right). Offspring that get no sex chromosome from their father and the XM from their mother are genetically female; those that get an XM from mom and XP from dad are genetically male (center). From “Sex chromosome transformation and the origin of a male-specific X chromosome in the creeping vole.” Couger MB et al. SCIENCE 07 MAY 2021:592-600.
How PSC Helped
Many of the most popular laboratory DNA sequencing methods can only sequence roughly 75 to 400 DNA bases at a time. The bases are the “A, G, T, C” alphabet of the genetic language. Because of that, scientists have to split the DNA into millions of small pieces in order to fully sequence them. If it weren’t for modern high performance computing (HPC), that would produce a huge mess of tiny snippets of DNA code with no way of putting them into the right order. But by matching the overlapping codes in these tiny fragments at light speed, an HPC system can put these millions of puzzle pieces together the right way, revealing the full code.
Once they had the sequence data from the lab, the collaborators began their computational work on the local computers and campus computing clusters available to them. But assembling the full DNA of the creeping vole’s sex chromosomes would have taken an unreasonable amount of time without access to computing systems with massive memory. The same as RAM on a personal computer, memory enables the system to keep more of the fragmented DNA sequences in “mind” at one time, enabling it to identify overlaps more quickly without repeated, time-wasting trips back to storage—the equivalent of a PC’s hard drive.

“This paper really required a lot of assemblies, which are really RAM-hungry for computational purposes. So I did a lot of the work on Bridges. It was really essential for me doing my work—the bioinformatics part [of the project]…If we didn’t have this wonderful public resource to run it on, the work wouldn’t have been possible.”—Brian Couger, Brigham and Women’s Hospital
Bridges, with its massive 12-terabyte extreme memory nodes, was the perfect match for this task. With about 1,000 times as much memory as a PC with good RAM, these nodes—of which the now-retired system had two—allowed the group to create an initial “short read” assembly relatively quickly. Using snippets of 100 DNA bases or fewer, such an assembly can use more easily generated data from the lab but also requires more memory.
What this assembly revealed was even stranger than Ohno had guessed in 1963. Again, it’s complicated.
Ohno was correct that females were XO. But instead of an X and a Y chromosome in males, the new results uncovered two strange types of X chromosome. The single XM chromosome the team found in both females and males looked mostly like a traditional X chromosome—except that it contained 10 genes only found in the Y chromosomes of other mammals. In males, the XM is paired with an XP chromosome that was even stranger—it was 90 percent “standard” mammalian X chromosome with a segment that represented about 10 percent of the total chromosome length with homology, or similarity in DNA code, to Y chromosomes.
In other mammals besides the creeping vole, having two X chromosomes poses a challenge for genetic females. If the genes on both copies were active at the same time, the X-chromosome genes would be twice as active as necessary. Instead, mammals have a mechanism called Xist that compensates for the two-gene dosage in females. In creeping voles it is the males who need help from the Xist mechanism—Xist silences the XP genes in most cells so that the males don’t have disruptive, double-strength gene expression from the two X chromosomes. Such gene silencing isn’t necessary in creeping vole females since they only have the single XM.
“Some features of this genome were so atypical that they were near unbelievable, such as conserved Y genes being present in females and a silenced somatic sex chromosome in males. However, other hallmarks of sex chromosomes in mammals, such as Xist expression for dosage compensation, was conserved even in this remarkable system.” —Brian Couger, Brigham and Women’s Hospital.
Stranger still, the male-only XP chromosome may have just seven of the 10 Y genes present on the XM chromosome. So it isn’t clear whether XP can fill the function of a Y chromosome or not (and is entirely silenced if it can’t), or whether XM and XP share the function somehow. Another question is how the females avoid activating those XM “Y genes” in their development. One thing is certain: it all works, because these rodents have so far survived for about four times longer than our own species.
The collaborators submitted a paper on their short-assembly sequence to Science, one of the most prestigious journals worldwide. The scientific reviewers at the journal were excited by the results, but felt that the short-read sequence didn’t quite nail down the evidence. That’s because short reads, starting with DNA fragments less than 100 bases, are more likely to contain matching errors. Because of these errors, the method can’t always put the DNA snippets in the right order. A more extensive, but harder-to-generate, “long-read” sequence, using lab-derived snippets of DNA sequence of more than 15,000 bases, can prevent this problem.
The authors of the paper agreed with the reviewers and went back to create a long-read assembly. Because the DNA sequences from the lab were now longer, the task required less memory but many CPU cores. So the new Bridges-2 platform’s 488 regular memory nodes, containing either 256 or 512 megabytes of RAM (still about 20 to 40 times as much each as the PC’s total RAM), were perfect for the job. In a marathon session over the 2020-2021 holiday season, Couger and his colleagues prepared a long-read sequence that upheld the results of the short-read and greatly improved the continuity of the assembly. The team’s work appeared on the cover of the May 6, 2021 issue of Science. You can find that article here.

