Combination of Bridges and Anton 2 Allows Team to Span Time Scales in Simulating Dagger-Like Microbe-Killing Molecule
Medical science is in a race to develop new and better antimicrobial agents to address infection and other human diseases. One promising example of such agents is the beta-defensins. These naturally occurring molecules stab microbes’ outer membranes, dagger-like, causing their contents to spill out. A scientist at Tennessee Tech University used PSC’s Bridges platform and the D. E. Shaw Research Anton 2 system hosted at PSC in a “one-two” simulation that shed light on a beta-defensin’s initial binding to a microbial membrane. The work promises clues to agents that can better destroy microbes with membranes.
Why It’s Important
Living in a pandemic world, it’s hardly necessary to point out how important new antimicrobial agents can be for treating afflictions from drug-resistant bacterial infections to COVID-19. One promising avenue of research focuses on the beta-defensins, a family of small protein-like peptides. These molecules, which consist of a chain of amino acids, are naturally produced by the body to kill bacteria. Better, since intact cell membranes are so fundamental to survival, bacteria can’t become resistant to this kind of attack. Beta-defensins can also suppress some viruses that have membranes, such as HIV. Engineered versions of beta-defensins may also be able to attack the SARS-CoV-2 virus that causes COVID-19. Scientists would like to know more about how the beta-defensins work. Such information could help them to design both new antimicrobial agents and drugs that help the natural versions of these peptides work better.
“My lab works on the defensins, which work by first crossing the microbial lipid membrane. By breaking the lipid membrane, they cause leaking of the microbial contents. Our goal is to understand defensins’ structure, their dynamics, and the functional relationship between them.”—Liqun Zhang, Tennessee Tech
Beta-defensins work like little daggers, stabbing their way into the membrane of a microbe and spilling its contents so it can no longer infect healthy cells. But these peptides work in a changing environment. The conditions in the body range from oxygen-rich oxidizing conditions surrounding cells, for example, in the lungs to oxygen-poor reducing conditions in cells in the intestines. This causes important changes in the folding of a beta-defensin peptide such as human beta-defensin 3. The amino-acid chain in this beta-defensin’s wild-type form is crosslinked to itself in three places via disulfide bonds. These links form in oxidizing conditions and break in reducing conditions. Scientists have long wondered how, and whether, beta-defensin can still destroy microbes in both forms.
To shed light on this question, Liqun Zhang at Tennessee Tech University found she needed to combine the complementary powers of PSC’s Bridges supercomputing platform and the D.E. Shaw Research (DESRES) Anton 2 supercomputer hosted at PSC.

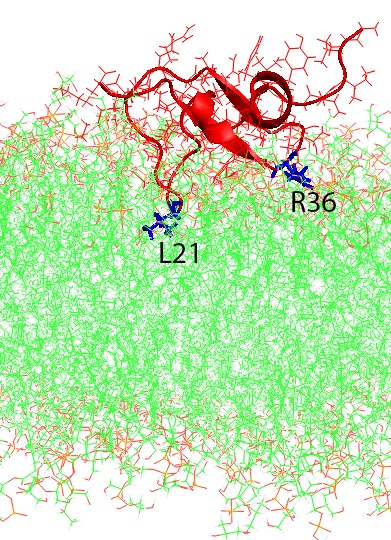
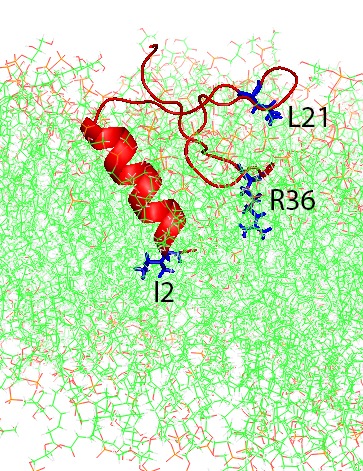
Molecular dynamics simulations of human beta-defensin type 3 (red and blue) in wildtype (left) and analog (right) forms binding to a simulated cell membrane (green). In both cases the two loops (R36 and L21) stick to the membrane, but in the analog form the “head” of the molecule (I2) sticks as well. Reprinted in part with permission from The Journal of Physical Chemistry B. © 2020, American Chemical Society.
How PSC Helped
As a first step, Zhang simulated the equilibrated form of human beta-defensin type 3. This consists of starting with the peptide’s chain in a disordered tangle and using the rules of chemical interaction to let that chain find the combination of twists and turns that it naturally settles into. Zhang found Bridges to be a great tool for this step. The National Science Foundation-funded platform’s massive computational abilities in both large memory and computational efficiency allowed her to simulate the initial 20 to 500 nanoseconds—billionths of a second—needed for the chain to find this lowest-energy form.
To simulate beta-defensin sticking to the membrane, though, she needed a much longer simulation—5 to 7.5 microseconds (millionths of a second), over 10 times longer. To perform this simulation, she used Anton 2, which is made available to PSC without cost by DESRES and supported through operational funding from the National Institutes of Health. Anton 2 is a highly specialized supercomputer designed and developed by DESRES that greatly accelerates such molecular dynamics simulations. Because of its specialized hardware and software, Anton 2 can perform up to 100 times as long a simulation in a given length of real time as a general-purpose supercomputer.
“The combination is necessary. Bridges can equilibrate a system for maybe 20 to 500 nanoseconds. Then we can move to Anton 2 for a long time span. We appreciate the computer resources that PSC offers; without their support, it’s hard for me to imagine how we could finish the work.”—Liqun Zhang, Tennessee Tech
Zhang simulated both the disulfide-crosslinked wild-type peptide and the uncrosslinked analog version of the peptide as it interacted with a virtual membrane typical of bacteria. In the initial Bridges simulations, she found that the wild-type version is much more rigid. Its crosslinks hold its shape more firmly than the analog version, which because of the lack of crosslinks is more flexible. The Anton 2 simulations showed an interesting difference that stems from this difference in flexibility. Two loops of the peptide chain initially stick to the membrane in both versions. But the analog version is flexible enough for an additional region, the “head” of the peptide, also to stick. Zhang reported her results in the Journal of Physical Chemistry in February, 2020.
It isn’t yet clear what the effects of this different way of initial binding to the membrane may mean for beta-defensin’s ability to destroy microbes. An important next step will be for Zhang to simulate the actual insertion of the peptide into the membrane and the disruption of the membrane. Another important step will be for Zhang’s colleagues to test her predictions on real peptides in the lab, verifying the results and in turn uncovering details she can use to create better simulations. Ultimately, she hopes that these simulations will offer clues for designing drugs to combat microbes that cause disease. Zhang and her colleagues also plan to design beta-defensin-based small antimicrobial peptides to combat Coronavirus.
You can read Zhang’s Journal of Physical Chemistry article here.