College Students learn molecular simulation on bridges-2
Educational allocation gives students taste of computational field, tools for science careers
Advanced computing has transformed the sciences, particularly biology. But college students often get no experience in wielding the supercomputers used in modern scientific research. That can make for a difficult transition from college to more advanced study. A class offered at Westminster College in western Pennsylvania has given biology and chemistry students simulation time on PSC’s Bridges-2 system, giving them tools to use later in their education. It also gave them a taste of the field that led many to choose career tracks involving computational biology.
Why It’s Important
While advanced computing has opened up powerful new ways of investigation in all of the sciences, it’s also posed a challenge for students. Today, you can’t study to be “just” a biologist or a chemist or even a social scientist and ignore computation. In virtually all those fields, computers loom large in identifying patterns too subtle or data too vast for the human mind to analyze.
Computing has particularly transformed biology. Biologists have been using computers to study the motions and functions of biomolecules for decades. Still, in recent years such molecular simulation has become a far more central part of the field. Even in the non-molecular fields, computation has become a bigger and bigger part of the scientific toolbox. It’s nearly true to say that, today, you can’t get hired as a biologist without at least a familiarity with computational methods.
“When I first started at Westminster, I had the idea that … students need to be exposed to computational chemistry, because it is becoming so prevalent in some way, shape or form in so many levels of research. So I implemented that … Last year was the first time I actually submitted an educational grant to the supercomputing center.”—Jessica Sarver, Westminster College
That’s why Jessica Sarver of Westminster College wanted to introduce her undergraduate students to molecular simulation. As part of a team-taught class that surveyed advanced scientific methods in molecular synthesis and analysis, she’d been offering instruction in molecular modeling using the students’ laptop computers. But the computational demands strained their machines, often crashing them. Sarver also wanted her students to get a taste of what’s possible with — and how to use — the kind of supercomputers that the pros employ to model the functions of the largest and most complicated biomolecules. To do this, she turned to the NSF-funded Bridges-2 advanced research computer at PSC.
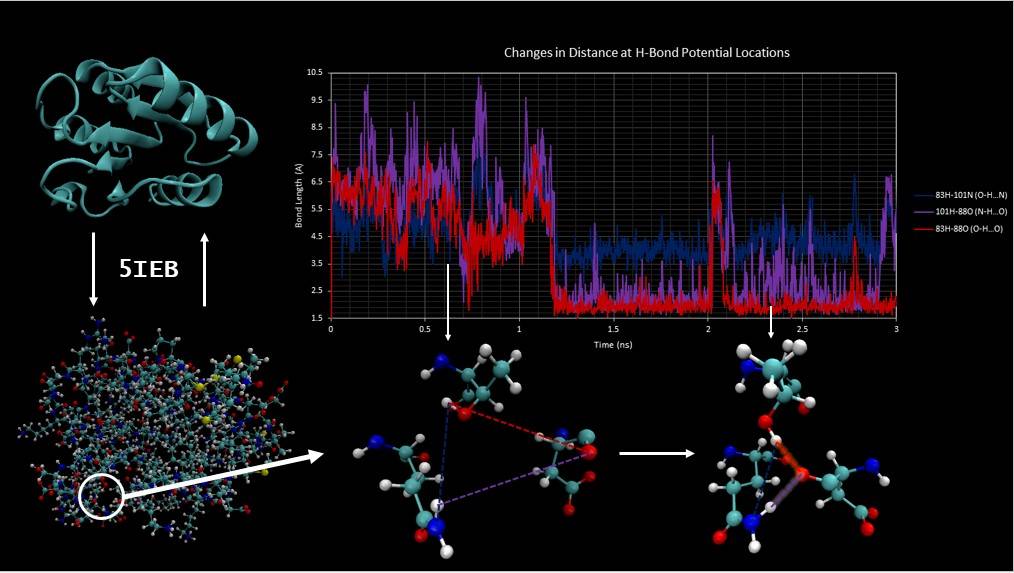
Above: Graphical abstract from Riana Smith’s project studying the serine-aspartate-repeat-containing protein G in the Westminster College Class. At upper left is the “ribbon structure” showing only the backbone of the protein, which is found in a bacterium that protects plants from infection. At lower left is the entire protein showing all of its atoms. Bottom center and right shows how bonds between hydrogen atoms and other atoms in the protein pulls part of the protein’s structure closer together. Arrows between the graph at upper right and the atoms below show time points for each of these structures.
How PSC Helped
Sarver herself had been introduced to computational chemistry as a biophysical chemistry graduate student. She had participated in “wet” laboratory work, using real-world experiments. But she’d also seen how a kind of ping-pong game between scientists using test tubes and scientists using computers could open up new questions for each to investigate. The lab results would give the computers data to make the models more accurate. The computational results would provide insights that the lab scientists could use to design new experiments.
Sarver wrote a successful proposal for an educational allocation on Bridges-2, giving her students computing time on the system. These students were not seasoned researchers, let alone expert computational scientists. She knew that they’d need to start simple, get a taste of simulating a molecule without necessarily delving deeply into its behavior. That way they could learn the basics of how to log onto the supercomputer, direct it appropriately and make sense of the results without getting overwhelmed. While she let them pick their own biomolecules to simulate based on their individual interests, the projects would focus on a very short simulated timespan — one to ten billionths of a second. This was long enough at least to see some movement in the molecules, if not enough to show major functional motions.
“The project that we did using molecular dynamics and simulations was our first-ever introduction to anything computational-chemistry based. Nobody [had] really used a supercomputer or high-powered computer at all, before taking that class … From that experience, I went on this past summer to do a summer internship [in computational chemistry] at the University of Wisconsin in Madison. [It] gave me the jumping point to further my career and to be able to perform different kinds of research.”—Riana Smith, Westminster College
Now in its second year, her supercomputing offering has proved a big success. The students’ backgrounds vary enough that each semester has been different. Most students found the class challenging, even intimidating, at first. But they also warmed to the work, eventually succeeding in producing good results. To ensure that the students were picking up more than a passing familiarity with the field, Sarver also taught them how to produce a “graphical abstract” that summed up their results in a visual way. She says that about half of the class’s combined chemistry and biochemistry majors go on to graduate school, where they likely encounter computational work in some way.
