PSC @ Supercomputing 2025
November 16-21 | St. Louis, MO
Booth # 311
We are excited to be in St. Louis for SC25! Our booth will have interactive demonstrations where you can explore how we use HPC to accelerate research.
PSC has been part of the national HPC community since 1986, and we are proud to continue our tradition of building relationships, fostering community, and of course, geeking out about research.
Kick Back at the PSC Backyard Party
Join us on Wednesday, November 19 from 4-6pm CST in Booth 311 on the exhibit floor. Enjoy a drink, unwind, network—and maybe play some giant Jenga.
Research on Tap
Be sure to mark your calendars and join us for our special events.

HuBMAP Demo: The Human Biomolecular Atlas Program
Booth 311
All Conference
- HuBMAP is an NIH Common Fund project that is working to create an open, global atlas of the human body at the cellular level to accelerate understanding of the relationships between cell and tissue organization and function and human health.
- We will be demonstrating the Exploration User Interface (EUI). With this tool, you can explore a 3D rendering of mapped organs and cells developed by HuBMAP researchers.
Learn about HPN-SSH
Location: Booth 311
All Conference
- HPN-SSH is a free and open-source data movement tool based on SSH.
It allows for speeds up to 50x faster than SSH – speeds of 8Gbps are not uncommon. - HPN-SSH is free, fast, and secure, and works with existing installs of SSH.
- We are creating an open-source ecosystem for HPN-SSH which includes building a community of developers, researchers, and users of the tool.
- We are asking people to help us with our market research by completing short surveys about membership options and pricing, to help us gain a better understanding of how to best serve the HPN-SSH community and future members.
Come Learn with Us!
Location: Booth 311
All Conference
- PSC’s Learning Lab is a collection of HPC self-paced, online courses.
- Ask for Valerie Rossi in the PSC Booth to talk more!
- Tell Us About Your Learning Path!
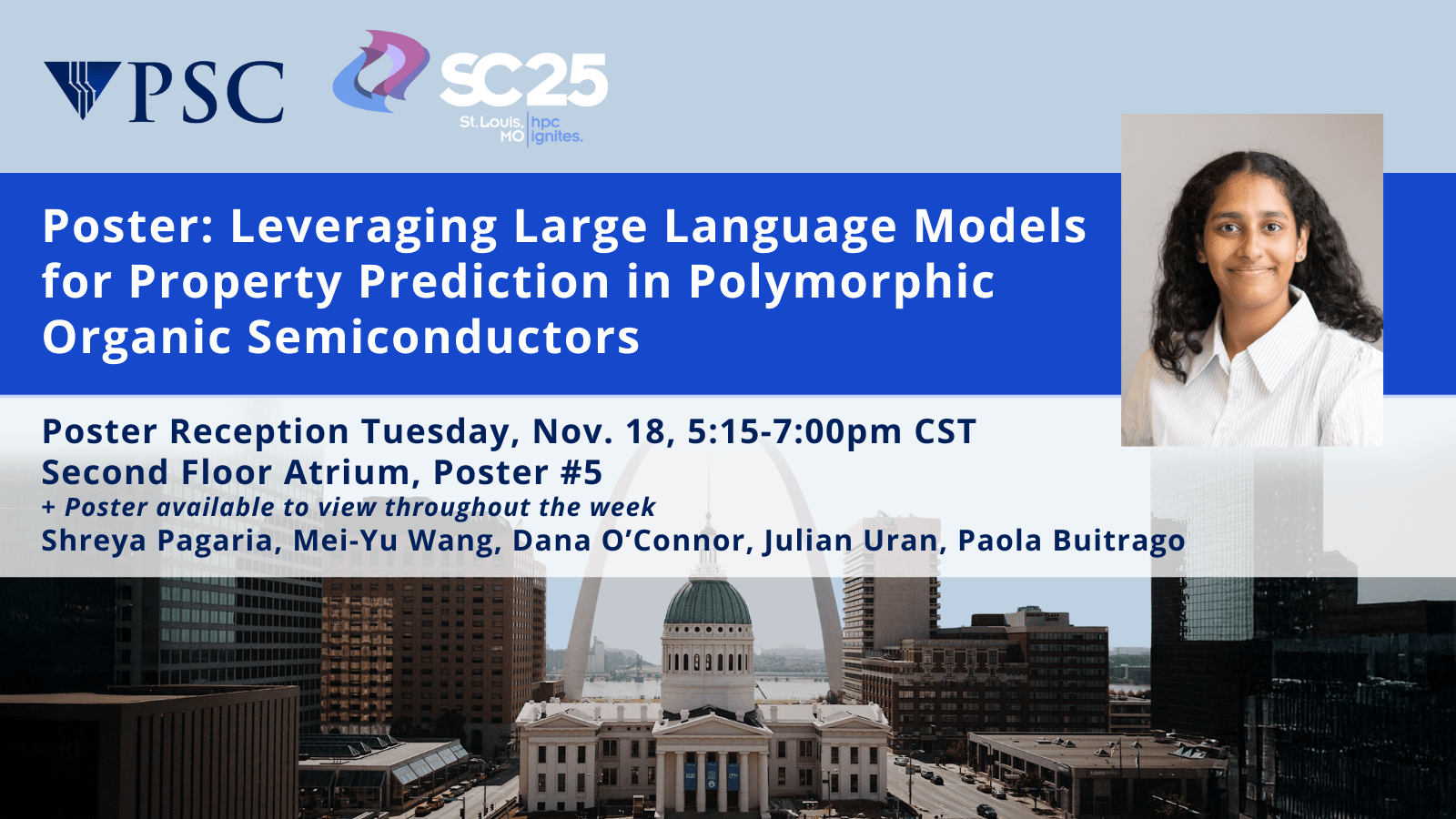
Poster: Leveraging Large Language Models for Property Prediction in Polymorphic Organic Semiconductors
Location: 2nd Floor Atrium, Poster #5
Poster Reception: Tuesday, Nov. 18, 5:15 – 7:00pm CST
Poster available to view throughout the week
Shreya Pagaria, Mei-Yu Wang, Dana O’Connor (former staff), Julian Uran, Paola Buitrago
Organic semiconductors (OSCs) are promising for next-generation electronics, but polymorphism complicates accurate property prediction and makes traditional methods costly. We investigate transformer-based large language models (LLMs) for predicting energy gaps in polymorphic OSC crystals. A Pegasus-managed workflow is deployed across heterogeneous hardware (PSC Bridges-2 and Neocortex Cerebras CS-2) to evaluate three crystal text encodings: Materials String, SLICES, and SLICES-PLUS against a baseline XGBoost Regressor model. The results show that the LLM-analyzed Materials String achieves the highest accuracy, particularly in polymorph-rich datasets, outperforming other representations in both pretraining efficiency and downstream tasks, as well as the baseline XGBoost results. These findings highlight the potential of LLM-driven crystal encodings to accelerate materials discovery and enable the scalable, data-driven design of organic semiconductors. Learn more.

PhySiViT: A Physics Simulation Vision Transformer
Location: 2nd Floor Atrium, Poster #89
Poster Reception: Tuesday, Nov. 18, 5:15 – 7:00pm CST
Poster available to view throughout the week
Mei-Yu Wang
Modern scientific computing generates massive simulation data across physics domains, yet researchers lack general-purpose tools for efficient analysis. While vision transformers like CLIP and DINO have revolutionized natural image analysis, no equivalent exists for physics simulation data. This project trains a custom Vision Transformer on “The Well” dataset, a 15 TB collection of diverse physics simulations. Using only 7 million images (compared to >100 million for CLIP/DINOv2), we trained our physics foundation model in 22 hours on a single Cerebras CS-3 server. Despite reduced training scale, our model demonstrates competitive classification
performance while exceeding at physics-specific tasks: temporal forecasting (𝑅2 = 0.33 vs. DINOvs2’s 0.23) and physics clustering (silhouette score = 0.232 vs. DINOv2’s 0.195). This work
demonstrates that efficient, domain-focused foundation models can achieve better performance in specialized scientific domains. Learn more.
BoF: Building Resilient and Sustainable HPC Communities Across Continents
Location: 263-264
Wednesday, Nov. 19, 2025 | 12:15pm – 1:15pm CST
Barr von Oehsen
This Birds of a Feather (BoF) session convenes global leaders of high-performance computing (HPC) communities to identify common challenges and share strategies for building and sustaining regional and national HPC ecosystems. Co-hosted by the UK HPC-SIG and the US CASC, the session builds on a successful ISC2025 BoF and CASC’s 2025 position paper on RCD regional collaboration. Participants will exchange funding and governance models, explore cross-border partnerships, and co-develop ideas for an international HPC Communities Network and shared resource hub. The session fosters peer-to-peer learning and lays the groundwork for a collaborative publication and ongoing global engagement. Learn more.

BoF: Super(computing)heroes
Location: 261-262-265-266
Thursday, Nov. 20, 2025 | 12:15pm – 1:15pm CST
Paola Buitrago
Members of underrepresented groups often lack access to role models within their minority. The HPC community is still predominantly male, making it difficult for young women to find female “superheroes” to identify with. Such role models are crucial for career planning and guidance. This session aims to provide especially women with the opportunity to meet influential, well-recognized female HPC “superheroes” from academia, research labs, HPC centers and industry. Join us to be inspired and find relatable role models as we work together to build a more inclusive and connected HPC community. Learn more.
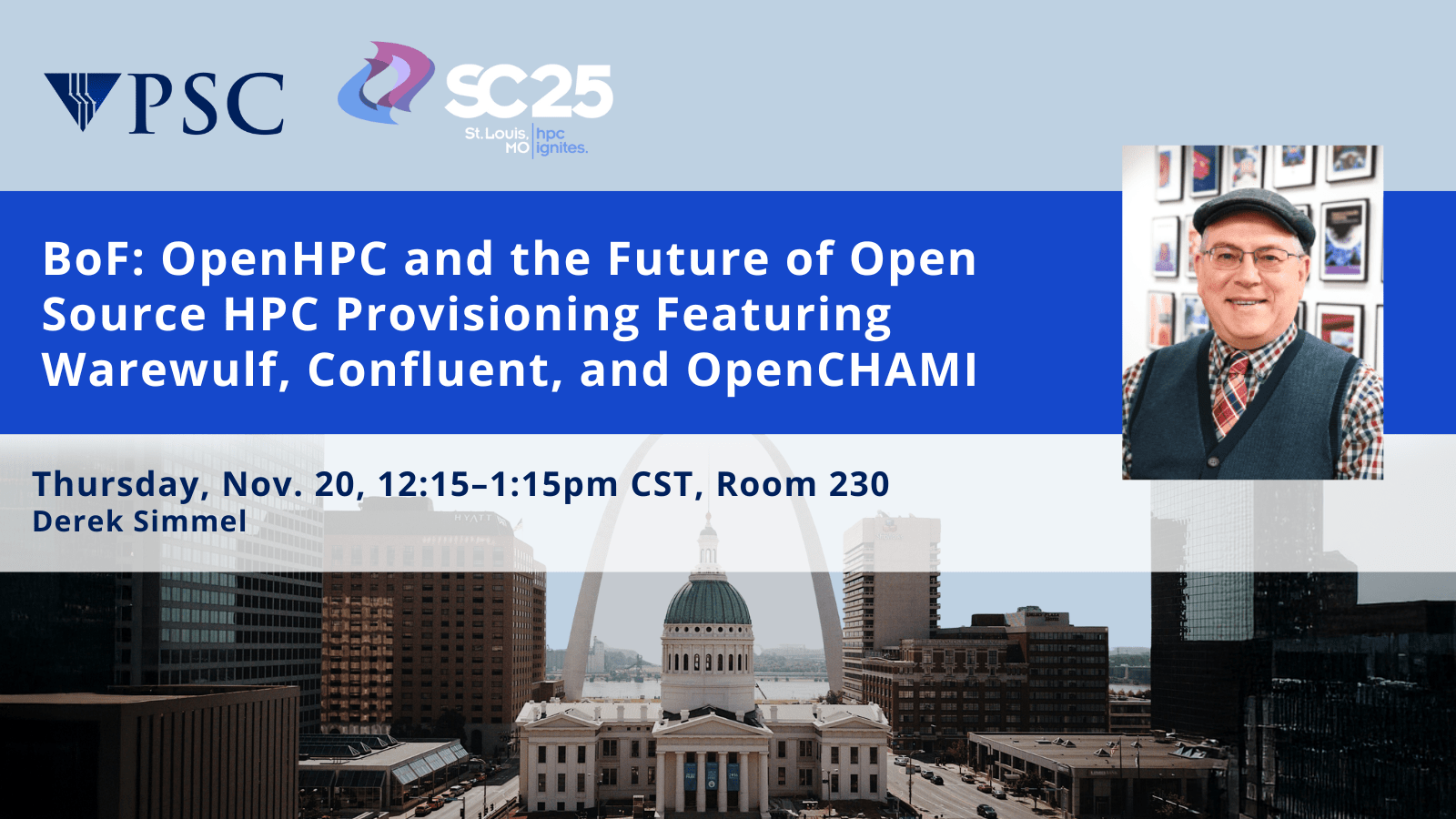
BoF: OpenHPC and the Future of Open Source HPC Provisioning Featuring Warewulf, Confluent, and OpenCHAMI
Location: 230
Thursday, Nov. 20, 2025 | 12:15pm – 1:15pm CST
The landscape of HPC cluster provisioning is rapidly evolving, with innovative open-source solutions emerging to meet modern computational demands. This BoF showcases leading open-source provisioning platforms through lightning talks from the Warewulf, Confluent, and OpenCHAMI communities, preceded by an update on the OpenHPC project. The session will highlight recent advances in container-based provisioning, cloud-native HPC management, and security-focused deployment strategies. Community members will engage in interactive discussions about best practices, interoperability challenges, and future collaboration opportunities. This forum aims to strengthen the open-source provisioning ecosystem and foster cross-project innovation for next-generation HPC infrastructure. Learn more.
About PSC
Pittsburgh Supercomputing Center is a joint computational research center with Carnegie Mellon University and the University of Pittsburgh. Our mission is to enable the advancement of science and research. We cultivate collaborative partnerships, empower the next generation of researchers, and provide cutting-edge cyberinfrastructure.
Apply to join our team today!
Join our team and help advance state of the art high performance computing, communications, and data analytics.
Pittsburgh Supercomputing Center (PSC) is a joint effort of Carnegie Mellon University and the University of Pittsburgh. PSC is located minutes from the heart of Pittsburgh, surrounded by culture and education.